Duration of the activity: 30 minutes
Recommended age: 15 years old and above
Population geneticsMedical Data Science
Hunt for variants
- Your turn to play
A new variant is here!
Find the differences, their impact and retrace the evolution of the coronavirus.
Activity 1
Find the differences!
Every time SARS-CoV-2 infects human cells, millions of copies of the virus, and thus of its genetic material, are produced…and those copies are not always identical!
Here are 3 small pieces of the genetic material of SARS-CoV-2:
the original and those found in two different copies (variant 1 and variant 2), coming from two different cities.
| original | ggt | ttc | caa | ccc | act | aat | ggt | gtt | ggt | tac |
| variant 1 | ggt | ttc | caa | ccc | act | tat | ggt | gtt | ggt | tac |
| variant 2 | ggc | ttc | caa | ccc | act | aac | ggt | gtt | ggt | tac |
Question 1
How many differences are there?
Answer
There are 3 differences (in green):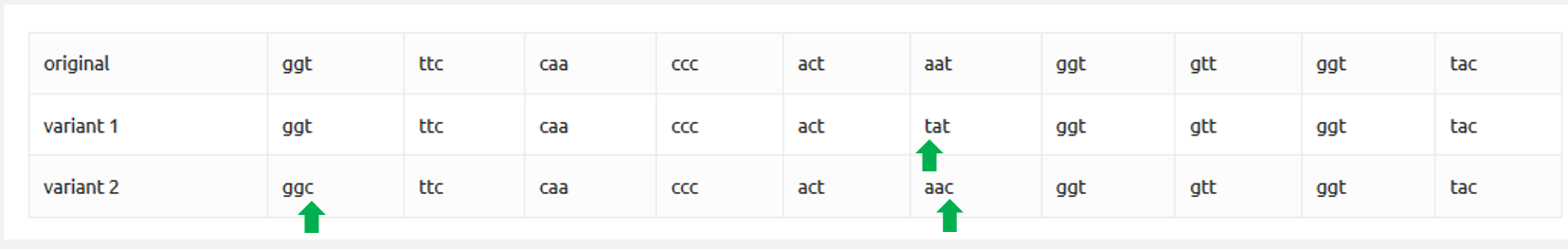
For those who are motivated…
Here is the alignment of the ‘original’ SARS-COV-2 genome with the genome of the variant Alpha :
Have fun counting the differences between these two sequences of 29’903 nucleotides…
Hint: The presence of a * means there is no difference, the presence of a – means a letter is missing.
Activity 2
One difference can make all the difference!
The genetic material of the virus contains all the recipes to make proteins. The differences (called mutations) found in the genetic material of virus variants can modify the proteins produced using these recipes…but not always.
One protein present at the surface of the virus, the spike protein, is particularly monitored by scientists. It’s this protein that enables the virus to enter human cells and it’s also this protein that is recognized by the antibodies we produce to fight the virus!
Depending on the differences, the virus could become more infectious or even escape the immune response provoked by a vaccine.
Translate the 3 nucleotide sequences into amino acid sequences using the genetic code:
add the missing amino acids in the sequence of the spike protein
The genetic code
3 nucleotide letters (A,T,C,G) correspond to a single amino acid letter (G,E,N,I,A, L, ...)
To translate, start from the centre and work outwards.
Example : GGT -> G
| POSITION | 498 | 497 | 498 | 499 | 500 | 501 | 502 | 503 | 504 | 505 |
| nucleotides (original) | ggt | ttc | caa | ccc | act | aat | ggt | ggt | ggt | tac |
| amino acids (original) | G | F | Q | P | T | N | G | V | G | Y |
| nucleotides (variant 1) | ggt | ttc | caa | ccc | act | tat | ggt | ggt | ggt | tac |
| amino acids (variant 1) | G | G | V | G | Y | |||||
| nucleotides (variant 2) | ggc | ttc | caa | ccc | act | aac | ggt | gtt | ggt | tac |
| amino acids (variant 2) | P | G | V | Y |
Question 2
How many differences are there between the protein sequences?
Compare it to the number of differences in Question 1…
Answer
There are 3 differences between the nucleotide sequences (in green) but only 1 difference between the amino acid sequences: N -> Y (in red).
A mutation in the nucleotide sequence does not necessarily modify the amino acid sequence of the corresponding protein, since the genetic code is ‘redundant’. Fortunately!
| …496 | 497 | 498 | 499 | 500 | 501 | 502 | 503 | 504 | 505… | |
| nucleotides (original) | ggt | ttc | caa | ccc | act | aat | ggt | gtt | ggt | tac |
| amino acids (original) | G | F | Q | P | T | N | G | V | G | Y |
| nucleotides (variant 1) | ggt | ttc | caa | ccc | act | tat | ggt | gtt | ggt | tac |
| amino acids (variant 1) | G | F | Q | P | T | Y | G | V | G | Y |
| nucleotides (variant 2) | ggc | ttc | caa | ccc | act | aac | ggt | gtt | ggt | tac |
| amino acids (variant 2) | G | F | Q | P | T | N | G | V | G | Y |
The mutation N->Y is found in the virus ‘variant 1’ at position 501 in the amino acid sequence of the spike protein: it is called N501Y.
This mutation could have an impact on the capacity of SARS-CoV-2 to infect human cells, as it is localised in the region of the spike protein that binds to human cells.
Activity 3
Track the evolution of the virus…
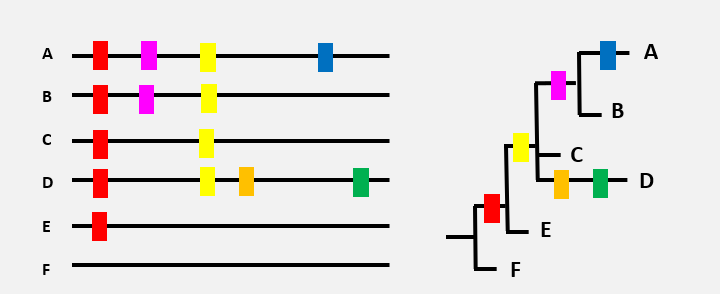
Here is a very schematized representation of the genetic material of SARS-CoV-2, each with a different combination of mutations (rectangles of different colours). Variant F is the sequence of the ‘original’ virus.
The order in which the different mutations appear can be represented by a tree. The red mutation, present in all the virus variants, probably appeared first (variant E), then the yellow mutation (variant C),…
Question 3
Which is the best solution?
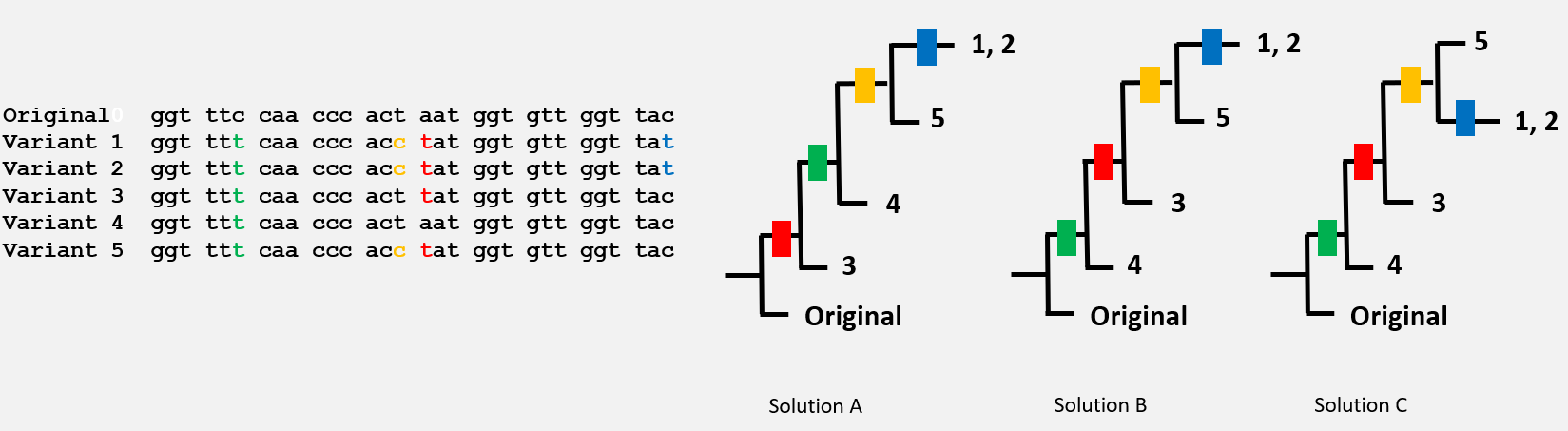
Here are the nucleotide sequences of 5 virus variants. Which tree showing the order of appearance of the mutations seems to be correct?
Answer
The correct answer is ‘Solution B’ and ‘Solution C’: these 2 trees are identical: the top branch, a bit like a mobile, has been inversed.
The variant 4 probably appeared before the others!
Activity 4
Here is a SARS-CoV-2 nucleotide sequence found in the wastewater in a treatment plant of a Swiss ski resort at the beginning of December 2020:
| position | 496 | … | … | … | … | … | … | … | … | 505 |
| codon | ggt | ttc | caa | ccc | act | tat | ggt | gtt | ggt | tac |
Is the mutation N501Y present? Could it possibly be the variant Alpha?
Hint: Use the genetic code and translate the nucleotide sequence…
Answer & Explanation
Here is the respective positions of the different codons and amino acids in the sequence of the spike protein:
| position | 496 | 497 | 498 | 499 | 500 | 501 | 502 | 503 | 504 | 505 |
| codon | ggt | ttc | caa | ccc | act | tat | ggt | gtt | ggt | tac |
| amino acids |
G | F | Q | P | T | Y | G | V | G | Y |
The protein sequence contains the amino acid Y at position 501, corresponding to the famous mutation N501Y.
For the anecdote, this mutation, as well as several other mutations present in the variant Alpha, were found in the wastewater from Lausanne and a Swiss ski resort. These samples had been collected between July and December 2020. There results suggest that variant Alpha could already have been circulating in Switzerland at the beginning of December 2020. This variant was officially discovered in Great Britain at the beginning of December 2020!
