Last update: February 2024
Population genetics Medical Data science
It makes a difference – Story
Genetic variation can sometimes have surprising effects...
Context
Blue or brown eyes? Blood type A or B? Do you think coriander tastes like soap? We’re all different, and the genetic basis of this individuality lies at the heart of our chromosomes, at the heart of our DNA.
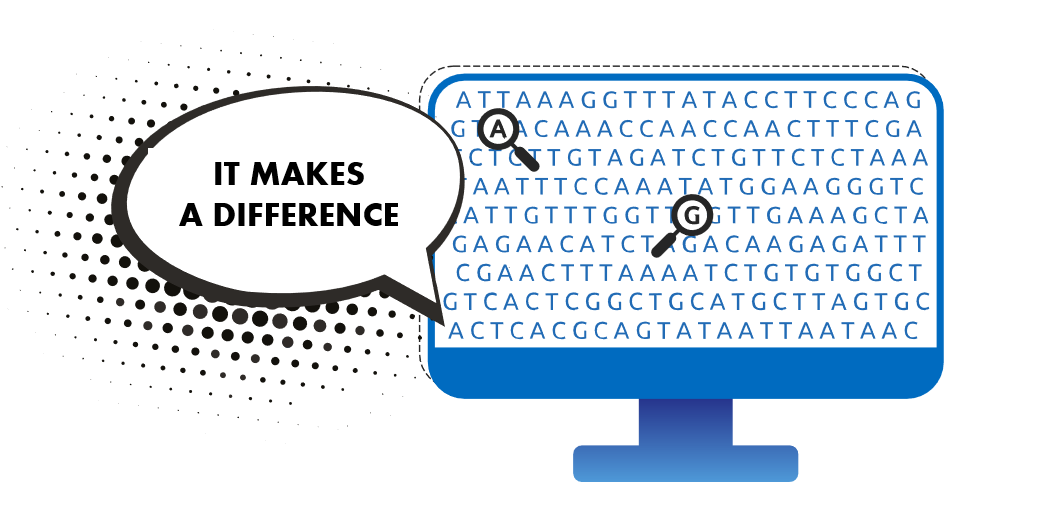
We have 23 pairs of chromosomes. A chromosome is made out of a long thread of DNA. DNA is made of a succession of 4 different nucleotides, known as a, t, g and c. The whole of our DNA – our genome – can be compared to a very long text made up of 3 billion a, c, g and t letters.
Here, for example, is the DNA sequence of chromosome 1.
Little changes in this text occur from one person to another. For example, a ‘g’ may replace a ‘t’ at a specific position. This is known as simple genetic variation, called Single Nucleotide Polymorphism (SNP). Around 1 nucleotide in 1000 differs between 2 individuals, i.e. approximately 3 million in total!
Researchers are trying to identify which of these 10 million or so known genetic variations are associated with a particular hereditary trait.
Many factors, including the environment, other genetic variations, and the location of the genetic variation in the genome, can influence its impact(s).
Some genetic variations can modify the quantity or ‘quality’ of a protein. These variations can have an impact on physiological characteristics such as eye color or resistance to a virus. They can also be the cause of genetic diseases or modulate the response to certain drugs. The majority of genetic variations have no consequences, and the impact of many genetic variations (or combinations thereof) is still unknown!
Why do some genetic variations spread more rapidly than others over generations? Researchers are asking a lot of questions, but finding the answers isn’t that simple!
Source: News from the Protein Mutability Landscape (2023)
Discover some examples of human characteristics that are influenced by simple genetic variations.
Click on “Learn more” to find out more!
Dry or wet earwax?
Earwax helps clean and lubricate the ear canal, and offers protection against bacteria, fungi, dust and water.
Genetic variations contribute to the type of earwax in the human population: wet or dry.
Blue is rare
Eye color is fascinating and depends on the presence of different pigments in the iris.
Genetics play an important role: some 150 genes are thought to be involved. And there are likely to be many many more as new studies are conducted!
SARS-CoV-2 resistance
Numerous genetic variations have been associated with susceptibilities to develop more or less severe forms of COVID-19.
We might have inherited some of these genetic variations from Neanderthal!
The acid taste
Acid taste is one of the five primary tastes, along with salty, sweet, bitter and umami.
The genetic mechanisms of acid taste detection have only recently been elucidated.

Alcohol intolerance
ADH proteins serve to break down alcohols, including ethanol, an alcohol found in alcoholic beverages or fermented foods.
These proteins are present in many species, and genetic variations can alter their function.
The speed gene?
The influence of genetics on sports performance is the subject of much study and debate.
Some 200 genes influence sports performance. And there are likely to be many many more as new studies are conducted!
ABO blood types
Blood types A, B, AB, and O were discovered in the early 20th century.
Why is there such diversity in the human population?

Beethoven's hair
In 1802, Beethoven requested that, after his death, information about his illness be described and made public.
His DNA revealed a few more secrets...

Dry or wet earwax?
Earwax helps clean and lubricate the ear canal, and offers protection against bacteria, fungi, dust and water.
Genetic variations contribute to the type of earwax in the human population: wet or dry.
Dry earwax is common in East Asians, while wet earwax is common in other populations.
A single g -> a genetic variation in the ABCC11 gene (located on chromosome 16) is responsible for determining earwax type. Individuals with ‘aa’ have dry earwax, and individuals with ‘ag’ or ‘gg’ have wet earwax.
The ‘g’ is the nucleotide that was present in our ancestors. The g -> a genetic variation is thought to have appeared in North-East Asia some 2000 generations ago, and then spread throughout the world.
A certain body odor
The smell of perspiration under the arms (known as armpit odor) is due to the action of bacteria present on the skin, which transform certain compounds produced by the glands that secrete sweat.
The ABCC11 protein is involved in the production of these compounds. The ‘aa’ genotype results in a loss of function of the ABCC11 protein, and an almost total absence of components typical of armpit sweat or wet earwax.
Caucasians and Africans have an active ABCC11 protein, wet earwax and a stronger axillary odor, while many individuals of Asian ancestry have an inactive ABCC11 protein, dry earwax and weaker axillary odor. These differences are of great interest to perfumers!
Why has dry ear wax been preserved over the course of evolution?
Mystery… here’s a guess:
Hypothesis – cold adaptation
The ancestral environment of East Asians would have been much colder than that of Africans. Ancestors of East Asians carrying the ‘aa’ genotype may have benefited from some advantage in having less abundant perspiration. This hypothesis does not rule out the possibility that other conditions, such as the microbial environment, may have played a role.
Source: A strong association of axillary osmidrosis with the wet earwax type determined by genotyping of the ABCC11 gene (2009) – A SNP in the ABCC11 gene is the determinant of human earwax type (2006) – The impact of natural selection on an ABCC11 SNP determining earwax type (2011)
Chromosome 16: ABCC11; rs17822931; UniProtKB: Q96J66
Blue is rare
Eye color is fascinating and depends on the presence of different pigments in the iris. Genetics play an important role: some 150 genes are thought to be involved. And there are likely to be many many more as new studies are conducted!
In 2008, researchers identified a genetic variation in the HERC2 gene (located on chromosome 15) that plays a significant role in eye color. They studied a large Danish family comprising a brown-eyed father, a blue-eyed mother, 17 children and 20 grandchildren.
An a -> g genetic variation in the HERC2 gene explains the blue color in the family. This variation leads to a lower production of the brown pigment melanin.
This same difference is found in the vast majority of blue-eyed individuals of European origin.
The history of blue
Our ancestors had brown eyes. The color blue is thought to have appeared in Europe between 6,000 and 10,000 years ago, probably in several stages, during the great migration of the first farmers from the Black Sea to Northern Europe.
Why have blue eyes stood the test of time and survived the millennia?
The question remains: eye color could be a neutral characteristic, with no significant impact on individual survival or reproduction.
Genetic variations in the HERC2 gene also influence skin color. The presence of the pigment melanin in the skin protects against UV rays. Its absence enables us to manufacture more vitamin D, particularly in regions with little sunlight.
Recently, researchers discovered a correlation between eye color and low-light vision. People with blue eyes would have better vision in low-light conditions than those with brown eyes. Further research is needed, however, to confirm a specific association between melanin content and low-light visual acuity.
The presence or absence of melanin in the iris has yet to reveal all its secrets!
Source: Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression (2008) / Effect of iris pigmentation of blue and brown eyed individuals with European ancestry on ability to see in low light conditions after a short-term dark adaption period (2024)
Chromosome 15: HERC2 rs12913832; several variations; UniProtKB: O95714
SARS-CoV-2 resistance
Numerous genetic variations have been associated with susceptibility to more or less severe forms of COVID-19. We might have inherited some of these genetic variations from Neanderthal!
A protective variation
The OAS1 gene (located on chromosome 12) codes for a protein involved in fighting viral infections (innate antiviral cellular response). Genetic variation g ->a has led to a less active and less abundant form of the protein. Today, over 60% of individuals have an ‘a’.
We might have inherited the ancestral form of OAS1 (with a ‘g’) from Neanderthal Man, over 40,000 years ago: it would have enabled our ancestors to resist several virus epidemics! And today, individuals with the ‘g’ variant are less likely to develop a severe form of COVID-19.
In other species…
Scientists examined animals known to harbor different coronaviruses. They found evidence of the active form of OAS1 in mice, cows and camels. Horseshoe bats, on the other hand, do not have the active form of OAS1. Horseshoe bats cannot prevent coronavirus replication in their bodies, and therefore could be reservoir hosts for certain coronaviruses such as SARS-CoV-2.
Variations that are not protective
We might have inherited from Neanderthal Man another genomic segment of around 50 kilobases, located on chromosome 3. This segment is associated with an increased risk of developing severe forms of COVID-19. This segment is present in around 50% of people in South Asia and around 16% of people in Europe. It is currently unknown which features of this segment confer a risk of severe COVID-19, and whether the effects of these features are specific to SARS-CoV-2 or to other pathogens.
Source: Host polymorphisms and COVID-19 infection (2022) – A prenylated dsRNA sensor protects against severe COVID-19 (2021) – The major genetic risk factor for severe COVID-19 is inherited from Neanderthals (2020)
Chromosome 12: OAS1 rs10774671; several variations; UniProtKB:P00973
Acid taste
Acidic taste is one of the five primary tastes, along with salty, sweet, bitter and umami. The genetic mechanisms of acid taste detection have only recently been elucidated.
The OTOP1 gene (located on chromosome 4) codes for a receptor protein that recognizes the molecules responsible for acid taste (citric acid (present in citrus fruits), lactic acid (present in fermented dairy products), acetic acid (present in vinegar), etc.). The OTOP1 protein is found in the taste buds of the epithelium of the tongue and palate.
Acid taste in history
Acidic taste is one of the five primary tastes, along with salty, sweet, bitter and umami. Dolphins seem to recognize only salty taste, while cats have no receptor for sweet taste. Surprisingly, the OTOP1 gene is found in all vertebrates. The first known vertebrates capable of perceiving acidic taste were ancient fish. This ability probably enabled them to test the acidity of the ocean and therefore the presence of carbon dioxide… Today’s vertebrates with the best acid taste perception are mammals, particularly primates.
Why has the ability to detect acidic taste been preserved throughout primate evolution?
Hypothesis 1 – vitamin C: Primates have lost the ability to produce vitamin C. It is therefore essential for these species to consume vitamin C. Recognizing acidic foods could be a way for them to ingest sufficient foods that contain vitamin C, such as citrus fruits.
Hypothesis 2 – fermented foods: the acidic compounds and alcohol produced during fermentation kill bacteria. So one way to find out if rotten fruit is edible is to test its acidity! It seems that ancient primates ate a lot of fermented foods.
Hypothesis 3 – another biological function: OTOP1 is the only gene known to encode an acid taste receptor. However, OTOP1 is also involved in the perception of gravity and acceleration in the inner ear, enabling balance to be maintained. Since the OTOP1 protein has another important biological function, changes in it could have had major consequences for the organism!
Source: The Cellular and Molecular Basis of Sour Taste (2022) – The evolution of sour taste (2022)
Chromosome 4: OTOP1; UniProtKB:Q7RTM1
Alcohol intolerance
ADH proteins are used to break down alcohols, including ethanol, an alcohol found in alcoholic beverages and fermented foods. These proteins are present in many species, and genetic variations can alter their function.
ADH proteins are involved in the breakdown of various alcohols, including ethanol. Ethanol is a toxic compound found in alcoholic beverages and fermented foods, but which we do not produce ourselves.
In humans, the ALDH2 protein plays an important role: it is responsible for the second step in ethanol degradation – the conversion of acetaldehyde, an equally toxic compound, into acetic acid, which is less harmful to the body.
Some 560 million individuals of Asian ancestry carry the same genetic variation (g -> a) in the ALDH2 gene (located on chromosome 12), which leads to a deficiency of the ALDH2 protein (ALDH2*2). Individuals who carry this genetic variation cannot tolerate drinking alcohol: they experience skin rashes, nausea, palpitations, and headaches as a result of the accumulation of acetaldehyde in their bodies.
This genetic variation is thought to have appeared in southeast China, ∼2,000 to 3,000 years ago. Why?
The ALDH2 protein is best known for detoxifying the alcohol we drink: there is little research on its potential role in other biological processes.
One hypothesis
Populations with a high prevalence of ALDH2*2 are most often located in areas where the hepatitis B virus is endemic. Infection with this virus and alcohol consumption have both an effect on the development of liver disease. The nausea and other manifestations associated with ALDH2*2 when alcohol is consumed could discourage alcohol consumption. ALDH2*2 individuals would therefore have been ‘protected’ during viral infections, resulting in positive selection of the ALDH2*2 genetic variation.
Source: Why can’t Chinese Han drink alcohol? Hepatitis B virus infection and the evolution of acetaldehyde dehydrogenase deficiency (2002)
The speed gene?
The influence of genetics on sports performance is the subject of much study and debate. Some 200 genes influence sports performance. And there are likely to be many many more as new studies are conducted!
One of the most famous genes in this field is ACTN3 (located on chromosome 11). It codes for a protein found mainly in fast-twitch muscle fibers.
A c -> t genetic variation in this gene, called ACTN3XX, results in a complete absence of the protein. This absence of ACTN3 appears to reduce the proportion of fast-twitch muscle fibers and increase the proportion of slow-twitch muscle fibers.
The ACTN3XX genetic variation is more frequently found in endurance athletes (e.g. cyclists and long-distance runners).
The presence of ACTN3 protein is associated with a higher quantity of fast-twitch fibers. The ACTN3 protein is most frequently found in athletes for whom strength or speed are important, such as short-distance sprinters. The ACTN3 gene is therefore sometimes referred to as ‘the speed gene’. The presence of the protein also seems to be associated with nocturnal teeth grinding!
Can an evolutionary loss become an advantage?
This genetic variation is thought to have appeared several hundred thousand years ago.
The loss of the ACTN3 protein would have been subject to positive selection, with its frequency increasing as man migrated from Africa to regions with a colder Eurasian climate.
The absence of the ACTN3 protein may have been beneficial in these living conditions. As a result, today around 1.5 billion people worldwide (18% of the population) are deficient in ACTN3.
Source: A gene for speed? The evolution and function of alpha-actinin-3 (2004) – ACTN3: More than Just a Gene for Speed (2017)
ABO blood types
Blood groups A, B, AB, and O were discovered in the early 20th century. Why is there so much diversity in the human population?
Blood groups are defined by the presence of certain sugars, called antigens, on the surface of red blood cells. The best-known blood group system is the ABO system, which is based on the presence of the A, B, and H antigens, associated with blood groups A, B, and O respectively. The presence of these antigens is the result of different combinations of genetic variations (alleles) that modify the activity of a protein that manufactures these sugars.
ABO in history
Most O alleles are derived from the A allele, so it’s possible that A and B alleles were more abundant at the start of human history. Today, the O allele is the most common worldwide (between 30% and 80% depending on the region).
A few hypotheses:
A, B and H antigens are known to play a major role in blood transfusion and transplantation, but they also have implications for human biology, as they are present in numerous tissues (platelets, intestines, respiratory system, etc.).
- The O allele is very common in regions of Africa where cases of malaria are frequent: the O allele could have a protective effect against malaria, a parasite that infects red blood cells.
- The presence of anti-A and anti-B antibodies in people with blood group O could protect against certain viruses, such as SARS-CoV-2.
- The A allele could protect against cholera, unlike the O allele.
- Several large-scale studies have shown statistical links between blood types and the risk of developing certain diseases (inflammation, cancer, etc.).
Much remains to be discovered. However, keeping the diversity of blood groups within the human population is undoubtedly an advantage from an evolutionary point of view.
Source: ABO blood group antigens and differential glycan expression: Perspective on the evolution of common human enzyme deficiencies (2022)
Chromosome 9: ABO rs1556058284; several variations; UniProtKB: P16442

Beethoven’s hair
In 1802, Beethoven requested that, after his death, information about his illness be described and made public. His DNA revealed a few more secrets…
In 2023, genetic analysis of DNA found in several strands of the composer’s hair indicated that liver disease and a viral infection may have been the cause of his death. The causes of his hearing loss and hair loss, however, remain a mystery.
Genetic diseases
Beethoven possessed a genetic variation in the PNPLA3 gene, a variation known to be associated with an increased risk of developing cirrhosis of the liver (non-alcoholic fatty liver disease 1).
He also possessed genetic variations in the HFE gene known to contribute to another liver disease linked to iron accumulation (Hemochromatosis 1).
A viral infection
The composer’s DNA also contained fragments of the hepatitis B virus. The researchers hypothesized that Beethoven suffered from a chronic viral infection that was reactivated in the months leading up to his death. This infection, combined with his alcohol consumption, aggravated his liver problems.
The composer’s genome was analyzed for genetic variations known to be linked to hearing loss or hair loss, but without success.
More secrets…
It was also found that he was 99% of European descent, probably lactose-tolerant… and that some of his family members were not legitimate children…
Source: Genomic analyses of hair from Ludwig van Beethoven (2023)
Chromosome 22: PNPLA3; UniProtKB:Q9NST1
Chromosome 6: HFE; UniProtKB:Q30201
Your turn to play
We are all genetically different. These small differences sometimes have surprising effects.
Discover a selection of genetic variations, their impact and their history, always in the light of evolution.
