Last update: December 2021
Population geneticsMedical Data Science
Hunt for variants - Story
Alpha, Beta, Gamma, Delta, Omicron... what’s that?
Discover SARS-CoV-2, its variants and their impact on the pandemic
Context
You know the story: a new coronavirus started to infect humans late in 2019.
Its name is SARS-CoV-2 (Severe Acute Respiratory Syndrome-related Coronavirus 2) and it is responsible for the disease COVID-19 (Coronavirus disease-2019).
This was the beginning of an incredible race against time to learn all about this virus and follow its evolution with the aim to control the pandemic, find treatments and vaccines.
What is a virus?
A virus is a very small parasite that can be compared to a bag containing genetic material. To multiply, a virus must infect the cells of a living organism (animal, plant or bacteria).
Life on earth would not be possible without viruses!
There are more viruses than stars in the universe. Every day, billions of viruses are deposited on a one meter-squared surface. Viruses play a key role in the equilibrium between populations and the evolution of species.
Source: Deposition rates of viruses and bacteria above the atmospheric boundary layer (2018); There are more viruses than stars in the universe. Why do only some infect us? (2020)
Viruses that make you sick
However, there are about two hundred types of viruses that cause disease in humans.
Source: ViralZone: Human viruses and associated pathologies
This is the case of the coronavirus SARS-CoV-2, which is responsible for COVID-19.
SARS-CoV-2...
SARS-CoV-2 infects human cells, and in particular the cells of our nose and our lungs.
The spike protein, present at its surface, can be compared to a key that allows the virus to enter these human cells.
Once inside the cell, hundreds of copies of the virus are produced thanks to the information present in its genetic material.
Every infected cell can then liberate several tens of thousands of viruses, some of which are ready to infect other cells! The infected cells are destroyed. In some people this can lead to serious breathing problems.
Source: The total number and mass of SARS-CoV-2 virions in an infected person
…its genetic material
The genetic material of SARS-CoV-2 is a single-stranded RNA. It can be compared to a thread composed of a long chain of nucleotides. There are 4 different nucleotides in RNA, symbolized by the letters A, U, C and G. Note that in the databases, RNA is always represented as DNA: each U is replaced by a T.
On January 10, 2020, Chinese researchers published the first sequence of the SARS-CoV-2 genome: it consists of 29'903 nucleotides.
Source: A new coronavirus associated with human respiratory disease in China (2020)

...its mutations
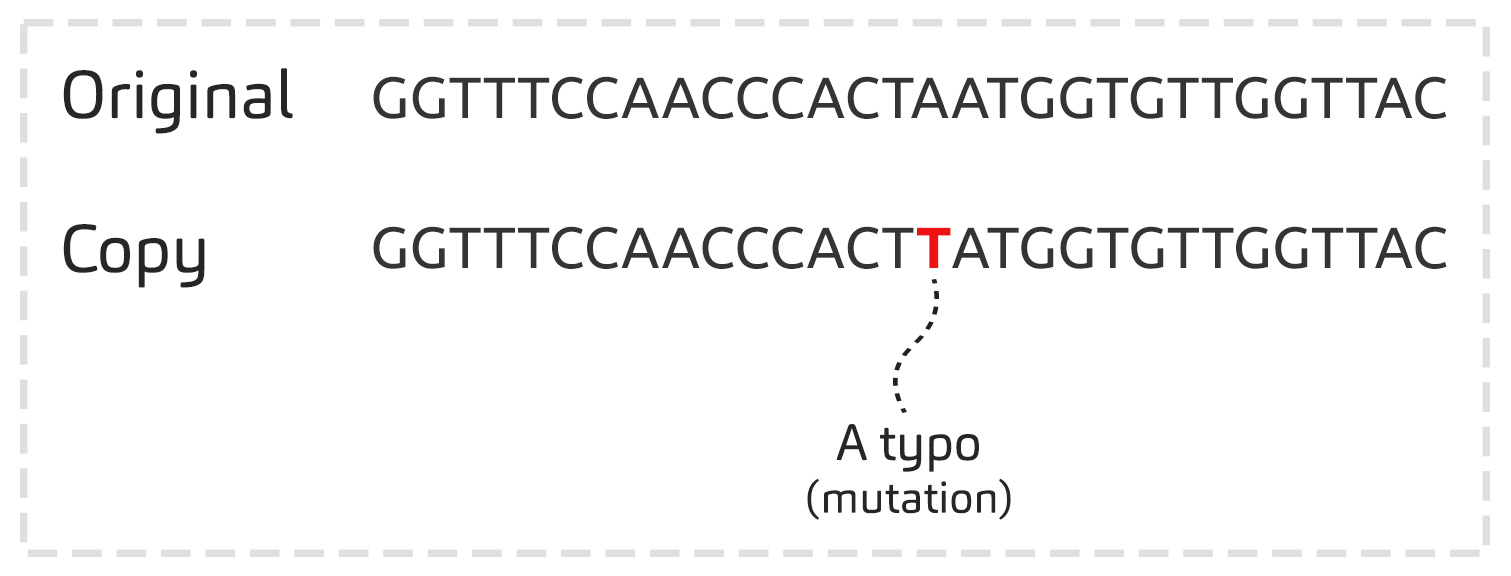
The genetic material of the virus is copied numerous times in the infected cells in order to produce thousands of new viruses.
These copies are not perfect: there can be up to 20 ‘typos’ per genome per year. These ‘typos’ are random: they are called mutations. Note that 20 ‘typos’ in about 30,000 nucleotides per year is a low substitution rate compared to other virus.
…and its variants
The viruses bearing certain mutations disappear with time, while others remain, because they are better adapted to their environment. The virus has to change to stick around. This, in a nutshell, is the principle of evolution!
Certain viruses then possess a combination of specific mutations that persist through time: these are virus ‘variants’.
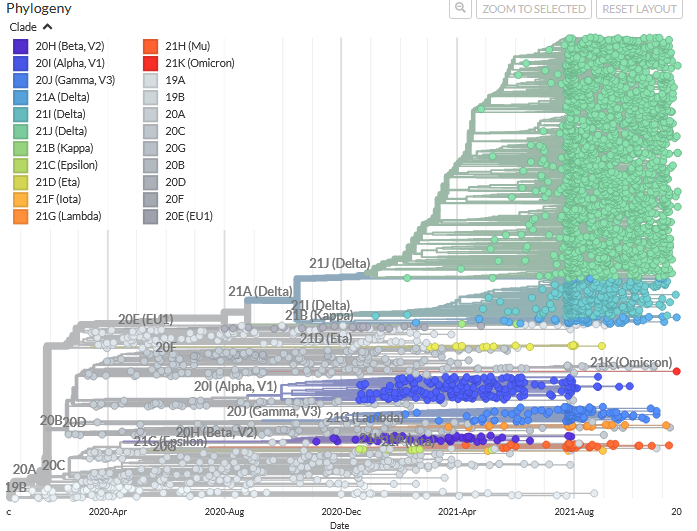
Today there are about twenty different virus variants: the most famous are Alpha (first detected in UK), Beta (first detected in South Africa), Gamma (first detected in Brazil), Delta (first detected in India), Mu (first detected in Colombia) and the latest one Omicron (first detected in Southern Africa in November 2021). There have even been variants that may have started in Switzerland!
Variants, mutations, some explanations
It is important to understand what is meant by ‘variants’ when referring to the evolution of SARS-CoV-2.
Each virus is slightly different, as virus contains a combination of different mutations. And most of these mutations have no influence on how fast the virus spreads or how sick it can make us.
The decision to regroup different SARS-CoV-2 viruses into some twenty variants is a bit arbitrary. The variants Alpha, Beta, Gamma, etc. are used to describe sub-families of SARS-CoV-2 comprising numerous samples, which certainly have mutations in common (but not all!) and which have a similar pathogenicity. They are a bit like talking about different dog breeds. In 2017, on the basis of genetic analyses, 161 dog breeds were regrouped into 23 groups or clades (groups of animals sharing a common ancestor).
Source: Genomic analyses reveal the influence of geographic origin, migration and hybridization on modern dog breed development (2017)
Identifying the circulating virus variants is essential to control the pandemic: certain mutations are clearly more dangerous than others!
Why are some mutations more dangerous than others?
The genetic material contains the recipes to make the proteins that are essential to the ‘life’ of the virus.
If a recipe is modified, because of mutations, the corresponding protein could also be modified.
Yet, some mutations modify the spike protein, which plays a key role in the pandemic!
The spike protein
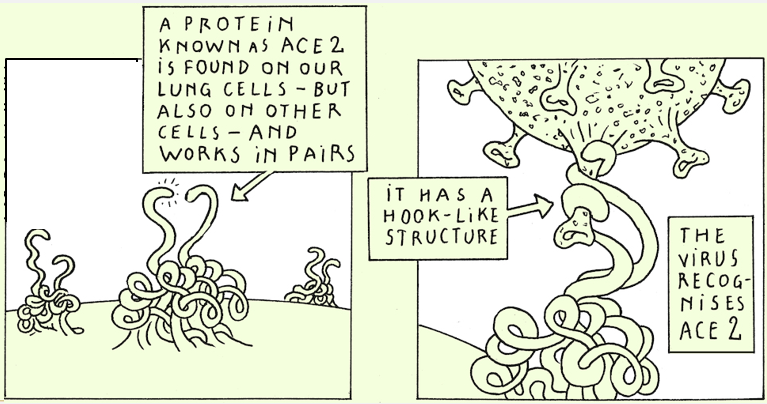
The spike protein is present at the surface of SARS-CoV-2. It’s this protein that enables the virus to penetrate in the cells by interacting with a human protein called ACE2.
It’s this spike protein that is recognized by the antibodies we produce, particularly upon vaccination. These antibodies are essential to fight the virus!
If mutations modify the spike protein, that could have consequences on the transmission of the virus and/or the efficacy of vaccines.
Spike under the magnifying glass
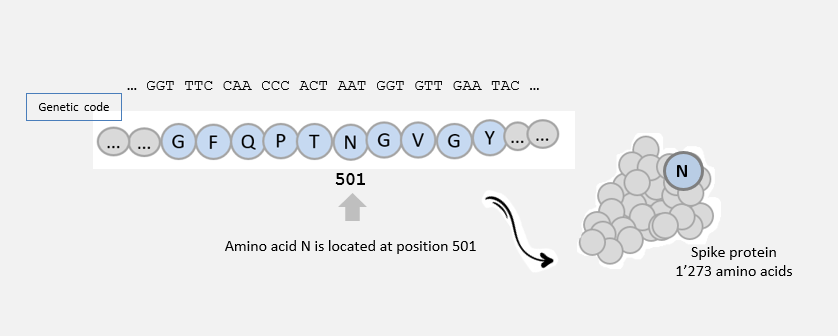
The spike protein, as all proteins, consists of a chain of amino acids. There are 20 amino acids, symbolized by letters (G, E, N, I, A, L, …).
The cell uses the information in the genetic material to make a protein.
3 nucleotide ‘letters’ correspond to 1 amino acid ‘letter’: for example, AAT -> N.
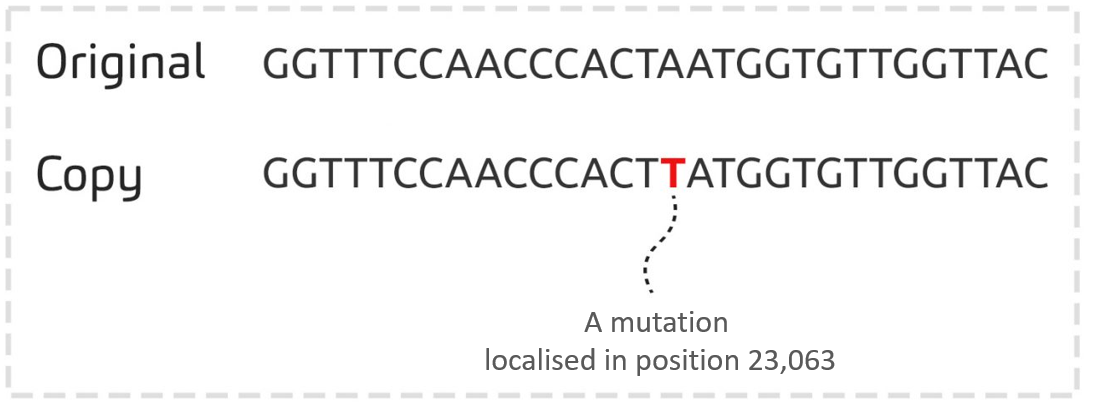
A famous mutation…
The mutation A -> T, localised in position 23,063 of the SARS-CoV-2 genome, is one of the mutations identified for the first time in a genome sequenced in UK in December 2020.
This mutation was then found in thousands of genomes from all over the world, and in particular in the genomes of variants Alpha, Beta, Gamma and Omicron.
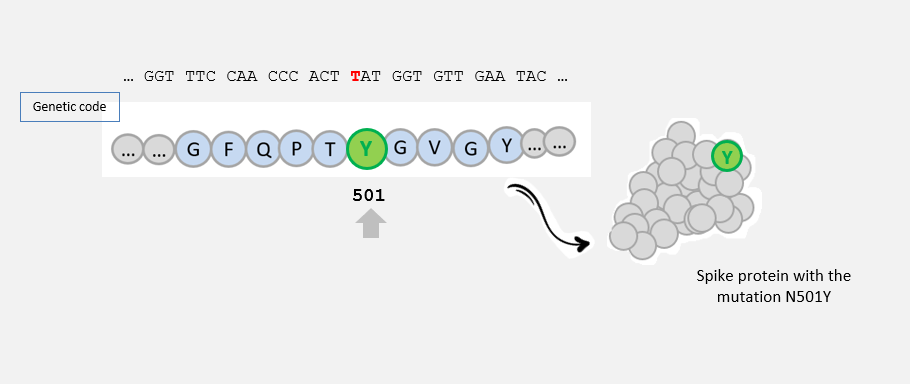
...N501Y
This mutation (AAT -> TAT) led to the amino acid change N -> Y at position 501 in the amino acid sequence of the spike protein: this mutation is called N501Y .
This mutation could have an impact on the capacity of SARS-CoV-2 to infect human cells, as it is located in the region of the spike protein that interacts with human cells.
Luckily, not all typos in the nucleotide sequence have an impact on the protein sequence!
Alpha, Beta, Gamma, Delta, Omicron, ...
The more a virus is allowed to replicated unchecked, the more chance it has to accumulate rare beneficial mutations.
Video: Coronavirus variants: What you need to know
The SARS-CoV-2 variants that persist through time have a combination of twenty or so mutations in their genome, compared to the genome sequenced in China in January 2020 (the reference genome).
Each virus variant has about ten mutations in the spike protein sequence compared to the 'reference' protein sequence (1'273 amino acids). The variant Omicron has about thirty mutations! (Source)
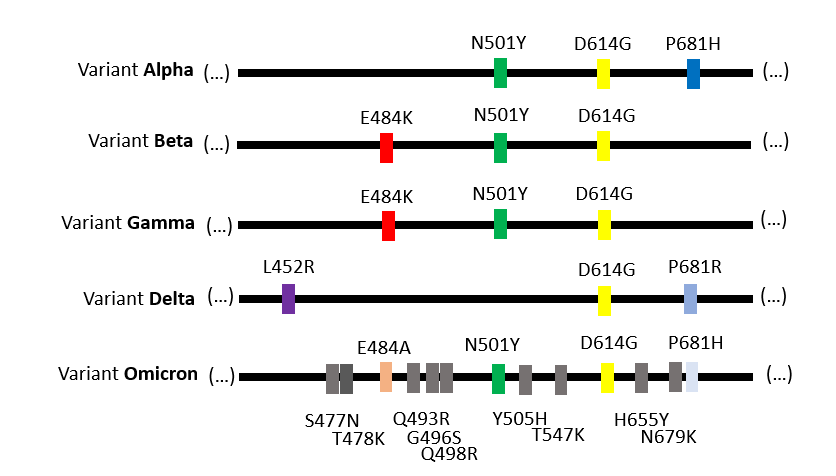
These combinations of mutations allow each virus variant to be identified, by analogy to a bar code.
For example, the mutation N501Y is found in the variants Alpha, Beta, Gamma and Omicron. It is not found in the variant Delta.
Each combination of mutations can have a different effect on the virus properties. That's why scientists work so hard to pinpoint the biological impact(s) of these combinations of mutations on infectivity, disease severity and vaccine effectivenes!
What does evolution have to do with all of this?
Sequencing the genetic material and finding the mutations allow the history of the virus to be reconstructed and hence to track its evolution.
Small puzzle
Here is a very schematic representation of 6 virus genomes (black lines), each with a different combination of mutations (differently coloured rectangles).
The genome of variant ‘F’ is the original genome.
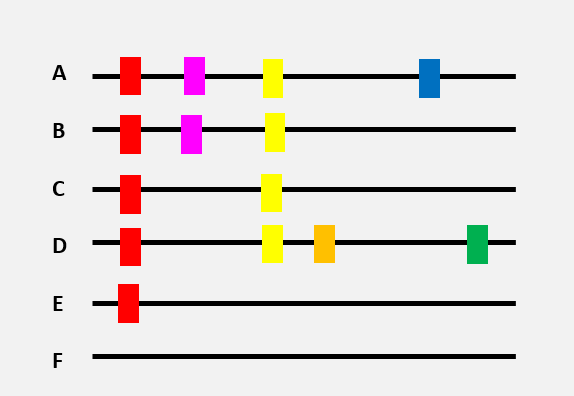
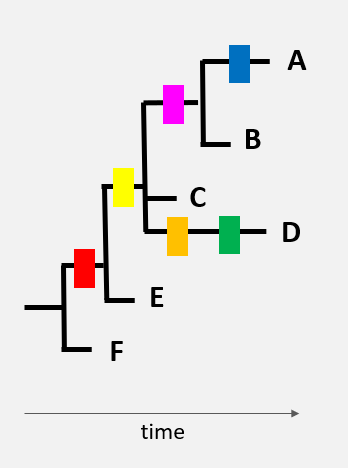
The appearance of different mutations can be represented by a tree. The red mutation found in numerous virus genomes was, without a doubt, the first to appear…
…then the yellow mutation
…then the pink, orange, and green mutations
One of the hypotheses that could be made, based on this tree, is that variant E was present before the other variants. By spreading in a population, the virus then acquired new mutations. New virus variants with specific combinations of mutations started to circulate (virus variant C and then A, B, and D).
By comparing the genomes in this manner, it is possible to reconstruct an approximate history of the transmission of the virus and its evolution.
Approximate, because we do not have access to all the genomes of all the SARS-CoV-2 circulating in the world, even though the experts compared several thousands of sequences every day in 2021!
Coronavirus and wastewater
Genomic surveillance is essential to detect the presence of different virus variants in the environment, to monitor their circulation and to estimate their abundance.
Researchers analyzed wastewater samples collected from several wastewater treatment plants in Switzerland between July 9 and December 21, 2020. DNA (and RNA) was extracted from these samples, sequenced and then analyzed: a real hunt for variants!
They found several mutations present in the variant Alpha in four samples from Lausanne and one sample from a Swiss ski resort.
These results suggest that the variant Alpha may have been circulating in Switzerland as early as mid-December 2020, i.e. two weeks before the official announcement of the first patient infected by the Alpha variant.